MBI Videos
Akira Horiguchi
-
 Madeline Edwards, Akira Horiguchi
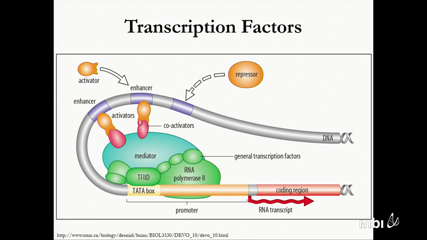
Madeline Edwards, Akira HoriguchiTranscription factors (TFs) often bind to specific DNA sequences to promote or block gene expression. The interactions between TFs and target DNA sequences may be regulated by DNA methylation. It has been well recognized that DNA methylation plays an important role in neural differentiation, which is determined by a cascade of TFs. However, the epigenetic regulated TF programs critical to brain development remain largely unexplored.
To fill in such a knowledge gap, we first analyzed mammalian brain methylomes to identify genomic loci differentially methylated during development. We compiled a set of experimentally validated TF binding sites from TRANSFAC 7.0, JASPAR 2014, and UniPROBE databases, and applied HOMER to identify TFs of which binding sites enriched in genomic regions hypomethylated in neurons and glial cells, respectively. Using MEME Suite and ClusterZ, we then determined pairs of TFs with binding sites frequently overlapped. With the Cytoscape program, we created a network of possible TF interactions, which can either be complex formation or regulatory. This predicted network provides novel insight to the epigenetic regulation controlling brain development.
